Published in the Science of the Total Environment journal, the study highlights the effectiveness and limitations of ozone treatment and invites further research into developing water-dependent purification methods, suggesting a more scientifically grounded approach to that aspect of fish farming.
“While ozonation is widely used at fish farms to purify water from organic compounds, no one has undertaken a detailed analysis of how this purification technique affects water quality and the levels of the various kinds of organic molecules,” study co-author senior research scientist Alexander Zherebker of Skoltech commented. “Using ultrahigh-resolution mass spectrometry we pinpointed the compounds susceptible to ozone treatment and their decomposition products. So, having these results fish farmers can draw conclusions as to whether ozone treatment is well-suited for their local water. At the same time scientists can follow up with complementary studies into other purification techniques. Ultimately, this can guide smarter business decisions based on modern science.”
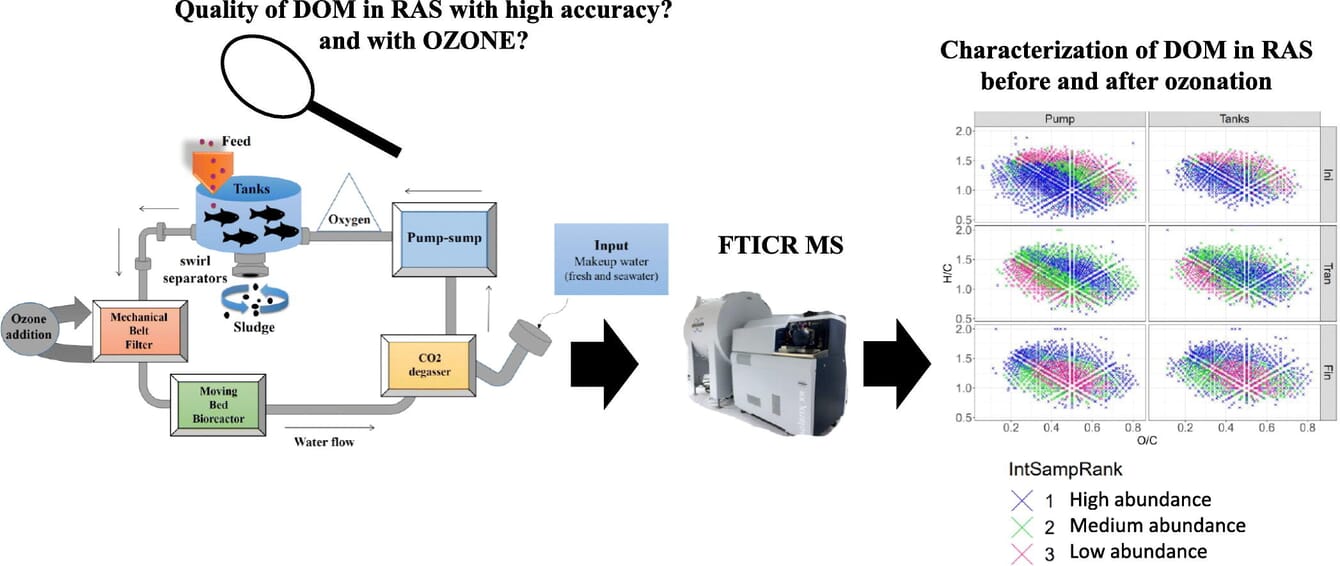
The organic matter that the researchers investigated enters the RAS tank from the natural water body that acts as the farm’s external supply source. The team repeatedly tested how the organic compound content and quality changed as time passed and as eventually ozone treatment was performed.
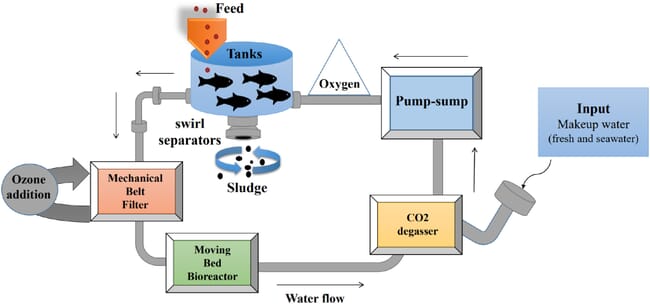
Experimental set-up of the recirculating aquaculture system (RAS) at the Nofima Centre for Recirculation in Aquaculture (NCRA).
“We see what kind of organics is coming in from the outside. Although it soon mixes with fish feed traces and waste products, we can clearly see the water starts out rich in humic-like and unsaturated CHO compounds. This stuff is associated with water blooming, changes in microelement transfer and microorganism activity; and obviously, it upsets fish. After ozonation the levels of these compounds drop and instead, we see greater concentrations of new fulvic-like and saturated compounds. Those are the decomposition products. They are more inert and therefore fish-friendly,” Zherebker explained.
Some compounds proved tolerant to ozonation, though — they may actually react with ozone but at higher doses. One caveat is that too much ozone will alter oxygen concentration and unavoidably impact fish health — they are only happy with natural oxygen concentration.
What enabled such a detailed analysis of dissolved organic matter was a highly sophisticated technique called Fourier transform ion cyclotron resonance mass spectrometry. It detects the presence and relative concentrations of various chemical species in a sample, without knowing exactly what you are looking for in advance — so-called untargeted analysis.

The researchers used Fourier transform ion cyclotron resonance mass spectrometry to identify different chemicals in the water sample © Freshwater Institute
Skoltech researchers, along with their Russian and Norwegian colleagues, ran mass spectrometry analysis and data treatment as part of the study. The Russian-Norwegian partnership was facilitated by the Transnational Access programme under the European Commission’s Horizon Europe funding programme.
“Our algorithm can now be applied to methods other than ozonation and to other systems to evaluate the effectiveness of different water purification techniques and select the best set of tools for a company’s individual needs,” study co-author Professor Evgeny Nikolaev concluded; he heads Skoltech’s Mass Spectrometry Laboratory, where the research was conducted.




