
Research fellow Andrew Coates and Professor Tim Dempster from the University of Melbourne on either side of project leader Nick Robinson, from Nofima © Fletcher Warren-Myers, University of Melbourne.
Salmon lice (L salmonis) can quickly adapt to new challenges, as the salmon aquaculture industry has discovered over the past decades. Many chemicals used to treat lice-infested salmon have become ineffective, due to lice evolving pesticide resistance. As new pesticides and other treatments become available to farms, it is crucial that the industry considers whether resistance could arise, and what actions farms can take to prevent this from happening. For this, a better understanding of how pesticide resistance grows and spreads across a network of salmon farms is required.
Researchers from the University of Melbourne, Nofima, the Institute of Marine Research and the University of Sydney have built a new computer model to simulate salmon lice evolutionary dynamics. This model is described in their new paper ‘A metapopulation model reveals connectivity-driven hotspots in treatment resistance evolution in a marine parasite’, which has recently been published in ICES Journal of Marine Science.
Modelling resistance
This model simulates lice infesting more than 500 salmon farm sites throughout southern Norway. It tracks how lice grow on farms, how they disperse between farm sites on ocean currents (using data from the Institute of Marine Research lice dispersal model), and how they evolve resistance to pesticide treatments. To test the model, the authors simulated lice adapting to azamethiphos – one of the chemical pesticides used on farms, to which resistance is now widespread. Their results match well with the current understanding of azamethiphos resistance: in just 10 years, the gene that provides resistance went from being very rare in the louse population to being very widespread.
The results highlight the link between how regularly farms were treated with azamethiphos, and how rapidly resistance evolved. The more frequently a farm was treated, the more likely that any lice carrying the resistant gene were to survive, breed and pass the gene onto the next generation. And as the population of lice on a farm became less susceptible to the chemical, treatments needed to be applied more frequently to keep infestations under control – which further accelerated evolution.
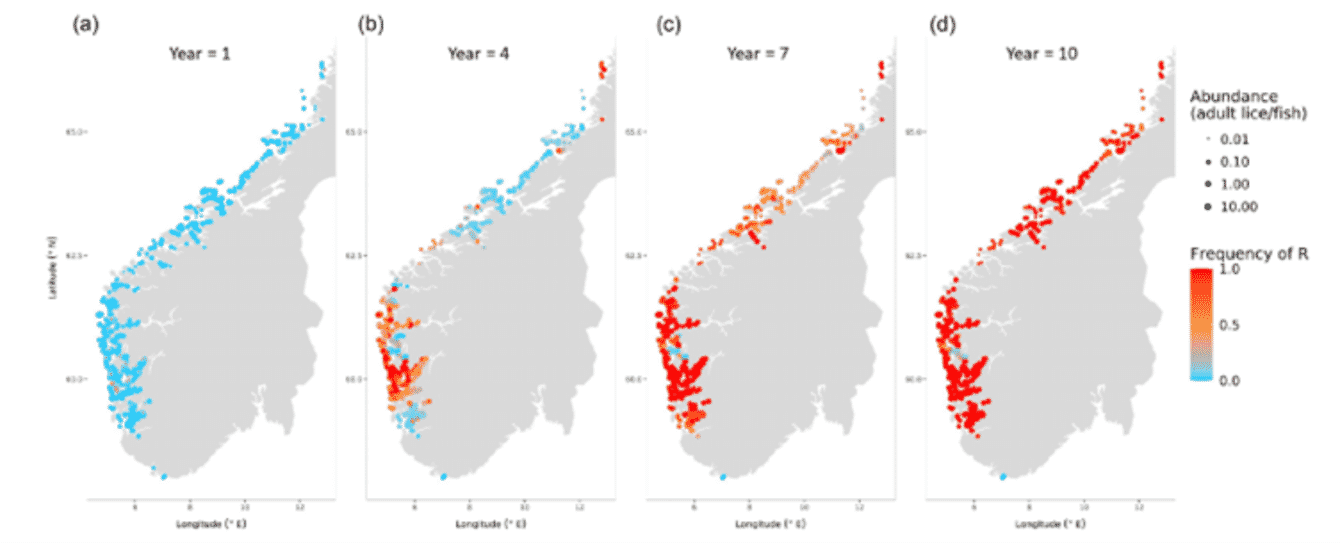
The colour of the points represents the frequency of the mutation (R) that gives lice azamethiphos resistance. Blue = most lice are susceptible to the pesticide; red = most lice are resistant. The size of the points indicates the abundance of lice on farms. Figure animation can be accessed at: https://cloudstor.aarnet.edu.a....
Evolutionary hotspots
Interestingly, there was a distinct spatial pattern to adaptation. That is, resistance evolved at different speeds in different parts of Norway. Resistance evolved most rapidly in the south-west region, around Hardangerfjord, before spreading northwards along the coast (Figure A). The authors describe this area as an ‘evolutionary hotspot’. By identifying evolutionary hotspots like these, researchers know which areas are most important when it comes to monitoring and managing for pesticide resistance.
Spatial patterns in how lice evolved resistance in the model were influenced by how likely it was that infective lice larvae were transmitted from one farm to another. Resistance evolved more rapidly in areas where farms were in close proximity and currents facilitated the transmission of lice infections from one farm to another. This was because high transmission of lice between farms led to high infestation rates and, in turn, more treatments. It also allowed for resistant genes to spread more easily to new areas. The evolutionary hotspot in Hardangerfjord is a region containing many farms in close proximity. As the salmon industry grows, the placement of new farms is critical. The model could be used to investigate how farm distribution could be optimised to reduce lice spread.
Alternative strategies to fight sea lice
The study also highlights the need for alternative methods of louse control that are more difficult for lice to adapt to than chemical pesticides. For example, the CrispResist project (funded by the Norwegian Seafood Research Fund) is investigating the potential for using gene editing to give Atlantic salmon high or full salmon lice resistance. The authors explain that their model can also help to decide on effective strategies to use for implementing these new technologies to limit the ability of lice to evolve and overcome the genetic resistance mechanisms introduced into the Atlantic salmon host population. Computer models can run countless different scenarios over large geographic areas and long timeframes that would be impossible to test experimentally. If lice can adapt to new management strategies (such as gene-edited salmon), then using models to predict the evolutionary responses of lice can find ways of slowing down, or even stopping, the spread of resistance.
The authors hope that this is the first of many uses of such models to better understand how lice respond to farm treatments at a regional scale. This knowledge can be integrated into farm management regimes, to ensure that new control technologies remain effective in the long-term.
The paper ‘A metapopulation model reveals connectivity-driven hotspots in treatment resistance evolution in a marine parasite’ can be accessed at: https://doi.org/10.1093/icesjms/fsac202


