Domestication is a natural process that occurs in organisms subjected to rearing in animal husbandry, horticulture and aquaculture, adapting them to artificial environmental conditions which differ from those their progenitor wild strains evolved within. Domestication is often coupled with selective breeding to further accentuate desired phenotypic traits such as enhanced growth rate.
Salmonids (salmon, trout and their relatives) provide an ideal model for domestication due to variability between individual fish for many desirable phenotypic traits, and, unlike for many domesticated agricultural species, in most cases wild strains remain as comparators to the domesticated strains. This difference in phenotype between wild and domesticated strains has been found by quantitative genetic studies to be determined mainly by additive genetic differences that accentuate phenotype in domesticated strains relative to the wild parental line. Our understanding of both undirected and directed selection occurring during domestication is based mainly on observations of the phenotypic characteristics of the animal (e.g. body size at maturation), with knowledge regarding the specific underlying genetic and physiological features under selection still poorly understood.
Improving our understanding of the molecular genetic basis of domestication will assist in identifying loci involved in control of phenotypic traits, both those desirable for the culture environment and those that may pose risks to wild strains. Indeed, of major concern to fish biologists is the intentional or unintentional release of domesticated fish stocks. Introgression of domesticated and wild genomes may result in hybrid progeny with reduced fitness for their environment. However, a recent study by Skaala et al. has demonstrated improved survival of hybrid progeny (some year classes) in the wild. To assist in addressing such concerns, it would be beneficial to understand genetic changes occurring during domestication as well as the effects of interacting genomes on gene regulatory processes. This knowledge would aid in the prediction of the detrimental outcomes of such crosses.
Inter-breeding between distinct populations can lead to different results depending on genetic and environmental factors. Hybridization of strains with low genetic variation and phenotypic range (e.g. from inbreeding or bottlenecks) may in theory yield hybrid progeny with desirable phenotypes through heterosis, where the offspring possess a more favourable phenotype than their parental strains.
Out-breeding depression can also occur where the off-spring express a non-favourable phenotype relative to parental strains, leading to reduced fitness of the hybrid progeny and the potential for negative impacts for wild stocks. Phenotype-genotype relationships have been examined for traits such as survival, aggression, predator evasion, and feeding motivation, and for growth potential of wild-domesticated hybrid progeny in relation to their parental strains. The overall findings of these studies suggested that domestication-induced traits are regulated mainly by additive genetic variation. Out-breeding with continually backcrossing of hybrid progeny into a wild genome may result in dilution of the domesticatedinduced phenotype and a reversion to wild phenotype.
While these studies have focused mainly on the effect of domestication through quantification of the phenotypic trait in the hybrid relative to both parents, the advent of microarray technology has enabled exploration at the mRNA level. Research on the relationship between mRNA levels and phenotypic traits has been reported in mouse and Drosophila sp. However, several studies have also applied this technology to explore the genetic variation arisen through domestication in rainbow trout, Atlantic salmon, coho, brook charr and lake whitefish salmonid species.
The present study uses microarray technology to screen for differences in mRNA abundance levels between fastgrowing pure domesticated, slow-growing pure wild, and wild-domesticated hybrid (intermediate growth) rainbow trout in liver tissue. Domesticated strains used within this study have been under selection for enhanced growth rate for greater than 30 years and show highly different growth rates relative to their wild comparators (leading to > 25-fold weight difference in domesticated compared to wild-type trout after 14 months), and are comparable to growth rates seen for growth hormone transgenic fish. The main aims of this study were to (1) investigate changes in mRNA levels that have arisen through domestication, and relate these findings to growth and other physiological changes, (2) investigate the relationship among genotypes regarding their effects on mRNA levels, specifically that for wilddomesticated hybrids relative to parental strains, and (3) investigate the effect of developmental stage on mRNA levels by inclusion of both age-matched and size-matched wildtype reference groups. Previous studies of domestication have tended to size and stage-match wild reference groups to domesticated populations in attempts to control for either developmental (body size, or stage) or environmental (rearing time) variance between groups due to size and stage differences. This study included both sizematched and age-matched wild groups as comparators.
Results
Entity list generation and statistical analysis
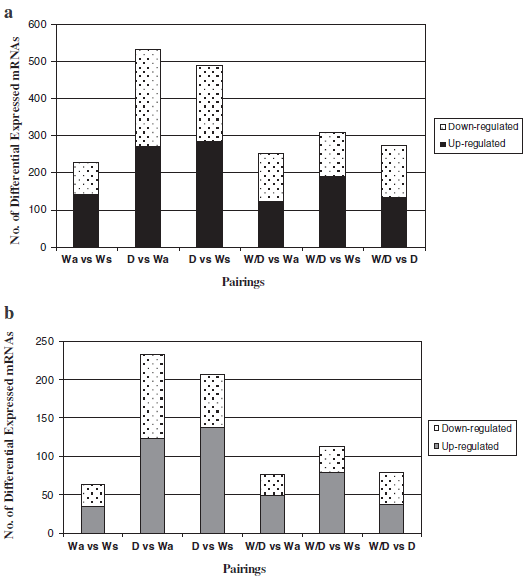
Levels of liver mRNAs were analyzed using a 44K oligoarray (consortium for Genomics Research on All Salmon Project; cGRASP) for four groups of rainbow trout (fastgrowing domesticated (D), wild-domesticated hybrid (W/ D), slow-growing age-matched wild-type (Wa), and slowgrowing size-matched wild-type (Ws)). Comparisons among these groups allowed assessment of the influence both of genotype (domesticated vs. wild, and hybrid) and developmental stage (body size and age) on mRNA levels. Following data normalisation and quality control filtering a total of 9,386 out of 43,689 oligos spotted on the array platform (21.5 per cent of those on the array) were found to be present within the confines of this experiment. Significant entity lists were generated for each group pairing and were also further filtered to examine differences greater than 2 fold to focus on mRNAs with major differences in expression. Figure 2 details the number of mRNAs found significant within each pairing, before (Figure 2a) and after (Figure 2b) filtering on fold change, and with replicate same named mRNAs entities removed. Statistical analysis using a one-way ANOVA was also performed with significance levels adjusted to P ≤ 0.01 for more stringent analysis (Additional file 7) which in most groups reduced the number of significant mRNAs to nearly half. In total, 733 or 7.8 per cent (same named replicates removed) of all entities expressed on the array were deemed to be significantly different (ANOVA, P ≤ 0.05) in terms of mRNA levels among any of the different rainbow trout group pairings (Figure 2a). mRNAs with major changes were defined as those having a greater than 2-fold difference (condition 1/condition 2) in mRNA level between any of the group pairings. This focus decreased the number of mRNAs to 351 (3.7 per cent) of all expressed entities on the array (Figure 2b) and will be referred to as the comprehensive significant entity list.
Differences in mRNA levels found for W (wild-type), D (Domesticated), and W/D (wild-domesticated hybrid) rainbow trout
Figure 2b details the number of mRNAs found to differ significantly among the various comparisons of D, W/D, Wa, and Ws trout. The largest proportion of mRNAs showing significant differences were noted for D when compared to W-type trout groups (greater than 200 mRNAs), with similar numbers found with comparisons to either age-matched or size-matched wild-type groups. Comparison of the W/D group relative to Wa and to D groups showed similar amounts of differential mRNA expression, whereas comparison of W/D to Ws showed higher amounts of mRNAs with different levels of expression. Some mRNAs (64) were also found to differ between the two wild-type groups (Wa and Ws).
Wa relative to Ws trout
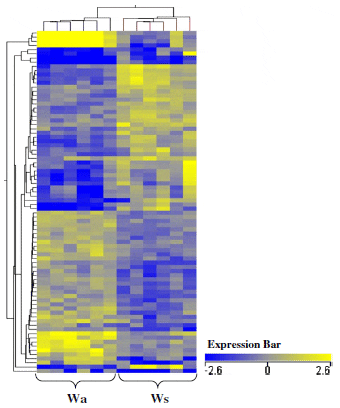
Differences in liver mRNA levels were noted between Wa relative to Ws-type trout (P ≤ 0.05). Of the comprehensive significant entity list, 18.2 per cent (64) differed in mRNA expression between Wa relative to Ws groups (Figure 2b). When visualized via heat maps of mRNA levels, individual mRNA expression patterns appeared consistent for all individual fish within each group but differed between the two wild-type groups (Figure 3). Expression profiles within this pairing show similar levels of up or down mRNA regulation (Figure 2b). Note that prior to the application of a fold change ≥ 2 (Figure 2a), Wa differed quite substantially in term of mRNA levels (229 mRNAs) in comparison to Ws-type trout.
D relative to Wa and Ws trout groups
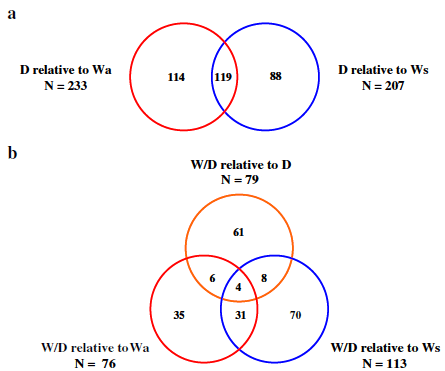
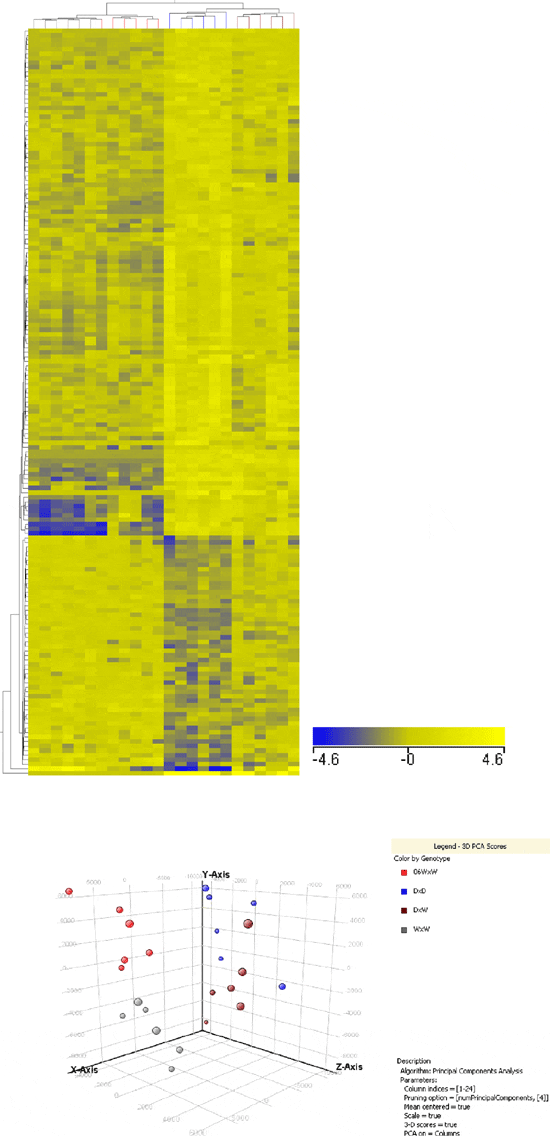
The largest proportion of differences in mRNA levels in liver was found in comparisons of D relative to either wild-type group. Of the comprehensive significant entity list, 233 (66.4 per cent) and 207 (58.9 per cent) mRNAs differed significantly between D and Wa and Ws, respectively (Figure 2b). Venn diagram analysis of both significant entity lists showed 119 of these mRNAs were shared in comparisons of D to Wa and to Ws-type trout (Figure 4a). Figure 2a and 2b detail the proportion of up and down mRNA regulation for all significant mRNAs found between pairings of D relative to W-type trout. For D relative to Wa similar levels of up and down mRNA regulation are noted, whereas for D relative to Ws a higher proportion of mRNAs were up-regulated as opposed to down-regulated (Figure 2b). Hierarchical clustering of those mRNAs found common to both wild-type in comparisons to D produced two cluster groups, with domesticated (D) and wild/domesticated (W/D) rainbow trout groups clustering together and, age-matched (Wa) and size-matched (Ws) wild-type rainbow trout clustering together (Figure 5a). Examination of mRNA expression levels (normalised expression value and fold change, see Additional files 1 and 2) and type (up or down regulation) were found to be very similar for both wild-type groups relative to D (Figure 5a). However, the unique differences in mRNA levels found here in comparison of D relative to Wa and D relative to Ws (Figure 4a) further emphasises the difference between wild reference groups. Individual variation (n = 24) within and between groups was examined using a 3D-PCA plot (Figure 5b). All samples within each genotype were found to cluster together within the PCA plot, with some overlap seen for D and W/D hybrids as expected.
W/D hybrid relative to D and wild-type trout
W/D hybrid group were compared to Wa, Ws and D groups. The largest degree of variation in mRNA levels was found in comparison of W/D with Ws, with 113 mRNAs (32.2 per cent) from the comprehensive significant entity list differing in this pairing. Similar proportions of differences in mRNA levels were found in comparison of W/D relative Wa and D groups (76 or 21.6 per cent, and 79 or 22.5 per cent, respectively; Figure 2b). A greater proportion of mRNAs were up-regulated in W/D relative to W-type fish, while similar levels of up/down regulation were noted for those mRNAs significant in W/D relative to D fish (Figure 2b). Venn diagram analysis of significant entity lists for each W/D pairing, show very few (n = 4) mRNAs to be common between all groups (Figure 4b). However, 31 mRNAs were shared between pairings of W/D to Wa and Ws-type trout (Figure 4b).
Relationship of genotype and mRNA levels
Those mRNAs (321) found to be statistically different (P ≤ 0.05 and fold change ≥ 2) between D relative to W-type rainbow trout during microarray analysis were used to determine the influence of genotype (domesticated vs. wild, and hybrid) and developmental stage on mRNA regulation. One-way ANOVA was perform on normalised log intensity values for D, W-type and W/D hybrid groups, to determine additive (a), D-recessive (r), or D-dominant (d) genotype effects for the 321 mRNAs (Table 1). Examples of each genotype effect are shown in Figure 6a. This list of differentially expressed mRNAs between D relative to W-type can be divided into three groups. Group A represents differentially expressed mRNAs found between D and both wild-type groups, group B represents mRNAs that differed significantly only between D relative to Wa type trout, and group C represent mRNAs that differed significantly only between D relative to Ws type trout. For group A, similar levels of additive, D-dominant and D-recessive genotype control were noted, however a tendency for increased prevalence of additive and D-recessive model can be seen (Table 1). Within this group it was found that a D-dominant genetic variation was more prominent for up-regulated as opposed to downregulated mRNAs (data not shown). Table 1 shows a large proportion of mRNAs within group A that were regulated in a D-recessive or D-dominant manner relative to domesticated fish. These results indicate a strong influence of both the wild and the domesticated genome in mRNA regulation. High levels of concordance (84 per cent of all mRNAs within group A), for additive, D-recessive and D-dominant effects, was found when using either Wa or Ws type trout for comparisons. This implies that for group A, mRNA regulation is most likely due to the effect of genotype and not a result of age or stage differences between D and wild fish.
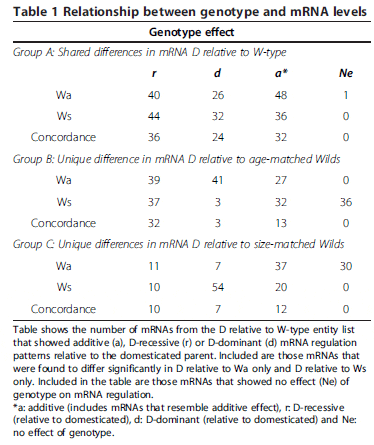
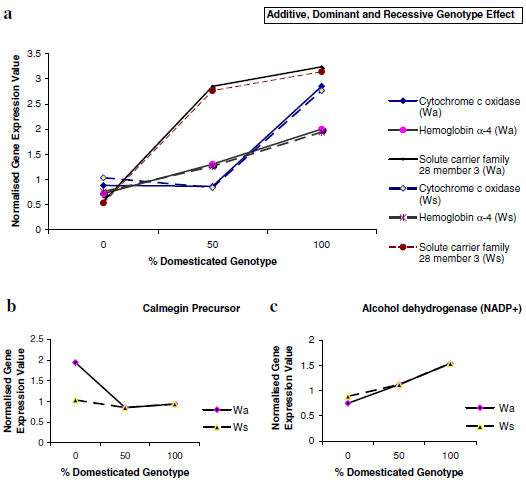
Table 1 shows that for groups B and C the greatest degree of concordance when analysing the effect of genotype (between Wa and Ws groups) on mRNAs abundance was seen for D-recessive regulation. A significant proportion of mRNAs within these groups were unaffected by genotype (Ne). For example, mRNAs that displayed a D-dominant response when comparing W/D to D and Wa groups, showed either no effect (Figure 6b) or were consistent with additive effects when comparing the same mRNA expression forW/D to D andWs trout groups. These results indicate that the controlling factor for differences in these mRNAs is most likely dictated by differences in fish developmental stage or life history between domesticated and wild rainbow trout rather than by genotype. Examples of additive (a), D-recessive (r) and D-dominant (d) genotype effects are shown in Figure 6a, along with illustrations of the effect of development stage on expression levels (Figure 6b) and those resembling additive (Figure 6c).
Physiological differences in liver for Wa, Ws, D and W/D rainbow trout
In order to determine which physiological pathways differ between the domesticated and wild-type strains examined, functional pathways were assigned where possible to the comprehensive significant entity list (351 mRNAs) identified from all group comparisons. The functional pathways were categorised under 16 umbrella terms (see Additional files 1, 2, 3, 4, 5, 6). Physiological pathways which showed alterations between groups were primarily associated with the following groups: response to stimulus (including stress/immune response and oxidation-reduction), cell/ tissue structure and development (including cell adhesion, muscle and cytoskeletal development), and transport (primarily in oxygen and metabolite transport).
Differences in physiological processes between Wa and Ws type trout groups were primarily in cell/tissue structure and development, response to stimulus, lipid metabolism, and transport (Figure 7a). In terms of expression profiles, it was noted that the majority of mRNAs which showed upregulation in Wa relative to Ws trout were associated with metabolic pathways, mainly lipid metabolism, transport systems and response to stimulus (oxidation-reduction reactions). Down-regulated mRNAs within this pairing related mostly to cell/tissue structure and development (skeletal muscle development) and response to stimulus (Figure 7a).
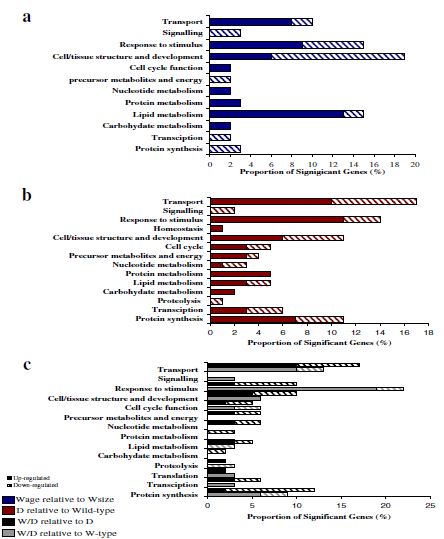
The main physiological pathways which differed between D to W-type groups were found in transport, response to stimulus, cell/tissue structure and development, metabolism (combined metabolic pathways) and protein synthesis (in Figure 7b only mRNAs commonly affected in Wa and Ws are shown). Of the mRNAs within this group, 15 per cent had unknown functions and therefore could not be assigned to any physiological pathway group. For mRNAs which demonstrated unique differences in mRNA levels in D relative to Wa or D relative to Ws, the proportion of mRNAs and types of physiological pathways were similar for both groups, but the specific mRNAs and mRNA expression profiles (up/ down regulation) differed (see Additional file 8). Some physiological pathways which differed in D relative to Wa, but not Ws, were homeostasis, protein metabolism, translation and apoptosis.
Figure 7c details the physiological pathways which differed between W/D hybrid relative to W-type and D-type trout (only shared mRNAs are shown for both W-types). Although few mRNAs were shared between W-type trout and D when either was compared to W/D (Figure 4b), some similarities were noted in the types of altered physiological pathways. While a large proportion of mRNAs within these pairings were unknowns, the main pathways identified were transport, response to stimulus, cell/tissue structure and development and protein synthesis. Difference in physiological pathways for W/D relative to D, in relation to W/D relative to W-type, were in signalling, generation of precursor metabolites and energy, nucleotide, protein and carbohydrate metabolism. Figure 4b details the number of mRNAs that differ in expression levels for W/D relative to Wa only and Ws only.
Discussion
To assess the genetic and physiological transformations that occur as part of the domestication process, the present study measured differences in mRNA levels between slowgrowing wild and fast-growing domesticated strains of rainbow trout, and analyzed the influence of combining domesticated and wild genomes in F1 wild-domesticated hybrid progeny. This research also assessed the effect of comparing mRNA levels of domesticated and hybrid genotypes to two different wild-type control groups that were either sizematched or age-matched to the domesticated genotype.
Substantial significant differences in mRNA expression were found for fast-growing domesticated rainbow trout relative to the slow-growing wild trout, with 5.4 per cent of all detected mRNAs differentially expressed between these genotypes. Further, significant differences were also observed for both of these parental strains relative to their wild-domesticated hybrid F1 progeny, although to a lesser extent than between parental groups. Among all detected mRNAs, hybrids possessed 3.3 per cent and 2.7 per cent of mRNAs with different expression levels relative to sizematched and age-matched wild trout (respectively), and 2.9 per cent of mRNAs differed when compared to domesticated trout. The lowest proportion of differentially expressed mRNAs was noted in comparison of agematched and size-matched wild rainbow trout, with 2.4 per cent of all detected mRNAs differing in this pairing. A related study comparing wild rainbow trout and a domesticated strain (different from the strain used in the present study) found similar results, where 6 per cent of all detected mRNA differed in liver tissue. Previous work on brook charr found 4.16 per cent of all detected liver mRNAs differed between domesticated and wild populations at the juvenile stage. Studies with Atlantic salmon whole fry found that 1.4 -1.6 per cent of all detected mRNAs differed in hybrid progeny with respect to their parental populations, whereas 6.4 per cent of all detected mRNAs differed in second generation farmed wild backcross relative to wilds. Similar results for hybrid relative to parental populations were observed in a study of normal and dwarf lake whitefish. Slight differences among these data sets likely arise by differences in experimental design (microarray platform, specific tissues vs. whole fry, developmental stages assessed) and variation due to the salmon species under investigation.
Similar proportions of mRNAs are up- vs. downregulated in domesticated fish relative to their wild counterparts (a slightly higher proportion of up-regulation was seen in the comparison with Ws as opposed to Wa trout). These results are different from the findings of Tymchuk et al. who found a higher representation of down-regulated mRNAs in the liver tissue of domesticated relative to size-matched wild rainbow trout. In the present study, many of the same mRNAs were found to be concordantly regulated in domesticated trout relative both to age-matched and size-matched wild trout. A concordant response is consistent with differences between slow-growing wild and fast-growing domesticated strains being changes that are stable across developmental stages and rearing conditions, and thus may be critical changes that have arisen during the domestication process. It is important to consider that although the domesticated strain used in this study has undergone selection for enhanced growth performance and shows vastly different growth rates to their wild counter parts, not all genetic differences between the strains will have arisen from domestication selection and not all will be related to growth. Other unintentional differences in behaviour, morphology and physiology likely have also arisen and could account for some of the genetic differences noted here. Between wild-domesticated hybrids and parental groups, proportionally more mRNAs were upregulated relative to age- and size-matched wild trout, whereas similar levels of up and down mRNA regulation were observed relative to domesticated trout.
The analysis of hybrid progeny in conjunction with the pure domesticated and wild parental strains allowed examination of the influence of genotype on mRNA regulation. Specifically, co-dominant expression is occurring if the level of mRNA product in the hybrid is intermediate between the parental strains, D-dominant expression if levels resemble the domesticated strain (equivalent to W-recessive with respect to the wild strain), D-recessive if levels resemble the wild-type strain (equivalent to W-dominant with respect to the wild strain), and over- or under-dominant if levels are respectively greater or lower than all the strains. For those mRNAs that showed concordant responses between domesticated and both wild-type groups (Wa and Ws), hybrid inheritance patterns showed similar levels of additive, D-dominant and D-recessive control. However, a slightly higher proportion of additive and D-recessive modes of regulation can be seen.
These findings are concordant with a previous study examining mRNA abundance differences between wild and domesticated strains of Atlantic salmon from two environments. The large proportion of mRNAs showing recessive and dominant genotype regulation suggests specific genetic influences of both the wild and domesticated genome in mRNA regulation in hybrids. The prevalence of dominant and non-additive responses reveals that introgression between domesticated and wild populations considerably alters the genetic control of mRNA levels from that evolved in wild individuals, and therefore may disrupt gene regulatory systems important for developing phenotypes for optimal fitness in nature. In contrast to the responses seen for mRNA levels, previous research examining the influence of genotype on selected traits such as growth and behaviour found mainly additive regulation. In such cases where F2 progeny have been examined, it is anticipated that the backcrossing of F1 hybrid genome combination to wild-type would further disrupt mRNA regulatory systems via outbreeding depression.
In contrast, numerous studies examining the influence of genotype on mRNA levels have found mainly nonadditive, dominant or transgressive modes of regulation. If we consider that the expression of a particular phenotypic trait such as growth rate or maturity are a result of complex networks of genes working in unison, as opposed to a single gene, it is possible that non-additive genotype effects noted in gene regulation responses are masked when gene complexes are formed to give rise to the visible phenotype. Based on the combined results of the current and many previous studies, we agree with the conclusion of Normandeau et al. and Roberge et al. that the consequences of hybridization on both mRNA regulation and phenotype expression are highly dependent on the specific genetic architecture of the crossed populations and therefore, highly unpredictable. The number of fish per genotype (n = 6) used in the present analysis of genotype effects on mRNAs regulation may have generated levels of variance that would prevent identification of differences among the groups for specific genes, and as such, actual differences may be somewhat greater. It is also possible that some of the effects seen may be a result of strain variation within each group which could generate non-homogeneity of genotype among the W/D hybrid fish selected, despite each showing intermediate growth rate and body size relative to the parental strains. Although the issue of within and between genotype variation was examined as part of this study and, little variation between individuals was noted, further study including analysis of additional individuals and strains would be beneficial.
To assess whether specific biological pathways were being influenced, functional assignments for differentially regulated mRNAs were determined for each of the genotypes. Differences between domesticated and wild rainbow trout showed a high representation of mRNAs involved in transport, metabolism, response to stimulus, cell/tissue structure and development, protein synthesis and transcription. The majority of mRNAs involved in these processes had higher mRNA levels in domesticated trout. Examination of alteration in fish physiology for domesticated and growth hormone (GH) transgenic coho and domesticated rainbow trout observed similar changes in stress and immune response, cell and tissue structure, energy production and protein synthesis physiological pathways. Comparable results for alterations in fish physiology have also been seen for brook charr and Atlantic salmon. The combination of these results suggests that similar biological pathways are altered in multiple species of domesticated fish, as well as GH transgenic fish, in order to support faster rates of growth, and that these changes may be both causal of or responsive to the underlying genetic variation that has led to altered phenotypes in fast-growing strains.
Specifically, in terms of metabolism, energy acquisition and utilisation, mRNAs displaying the highest level of fold change between domesticated and wild strains were Apo-A1-2 precursor, ELOVL FA elongase 6, FAA and protein canopy homolog 2 precursor, NADH-ubiquinonereductase chain 2, cytochrome C and COX IV-2, with under expression of ACBP. The elevated levels of many mRNAs involved in lipid metabolism may reflect the greater demand in domesticated fish for growth-related resources necessary to support increased growth performance. Additionally, many mRNAs associated with protein synthesis were overexpressed in domesticated trout. Protein synthesis plays a strong role in fish growth through mediation of many growth related pathways. mRNAs which displayed the highest level of over-expression were 40S ribosomal proteins (S5 and S21) and superoxide dismutase. Although superoxide dismutase has been linked to other roles which include antioxidant defence, it is included within this group for its role in protein biosynthetic process and growth regulation. Given that the liver tissue is highly involved in protein turnover, amino acid metabolism and lipid metabolism for its role in protein biosynethic during periods of accelerated growth the elevation of mRNAs within these pathways for the domesticated population was not unexpected.
During this study many mRNAs associated with oxygen and macromolecule transport were found to be expressed at a high level in domesticated relative to wild trout. mRNAs involved in these pathways may show increased levels due to the higher metabolic rate of the domesticated trout, required for increased growth and nutritional absorption. Notable with respect to transport, mRNAs involved in ion transport were found to be down-regulated in domesticated fish. mRNAs which displayed the highest level of over-expression were, solute carrier family 28 member 3, hemoglobin subunits beta (β and β-1,-2) and alpha (α and α-4) and vitellogin- 3 precursor. Similar results of increased expression of hemoglobin beta and alpha subunits in liver tissue were reported by Rise et al. for examination of GH transgenic coho expression. Tymchuk et al. observed different results to those described above, with increased expression of hemoglobin found only in the brain tissue of domesticated rainbow trout. Differences seen between studies maybe due to, strain selection, variance in fish growth rate, or, perhaps to differences in the vascular circulatory systems in the liver tissue.
Results of differential mRNAs expression in liver tissue showed over-expression in domesticated trout of many mRNAs involved in the activation and regulation of the complement pathway and innate immunity. Increased expression in domesticated trout was noted for lectin precursor, Complement C3-1, Fucolectin 6 precursor, Complement factor b precursor, C-type lectin domain family 4 member m and decreased expression of primary defence mechanisms exemplified by Ig mu chain C region membrane bound form. These results are contradictory to the findings of Tymchuk et al. who described down-regulation of many mRNAs involved in the stress and immune system, and attributed this fact to tradeoffs incurred by domesticated trout in order to sustain increased growth rates. Debes et al. also noted down-regulation of some immune related mRNAs in domesticated Atlantic salmon. However, up-regulation of CD59 and MHC class II combined with higher levels of lysozyme C transcript was also noted, and is different from the findings of Tymchuk et al. It was suggested that elevated levels of these mRNAs may relate to domesticated fish displaying a higher resistance towards vibriosis, a common bacterial disease in aquaculture relative to wild populations. It is possible that increased expression of mRNAs involved in the complement pathway may be explained by an increased host defence system in domesticated rainbow trout, an acute phase response prior to sampling, or alternative uses for these mRNAs in regulatory pathways in domesticated trout.
An objective of this study was to investigate changes in mRNA levels in domesticated trout using different wild-type comparators. Whenever two strains with different growth rates (e.g. wild and domesticated) are being compared, they will, at a specific age, naturally have different body sizes if provided with satiating levels of food. Since body size is linked to developmental stage in many fishes, differences in mRNA levels are anticipated between such groups simply due to the fish being developmentally distinct, rather than due directly to genetic differences causal of the domestication phenotype. To date, studies investigating the effect of wild reference group selection have focused mainly on wild populations from different geographical location, population groups, life stages and environmental treatment. Bougas et al. clearly demonstrated the influence of wild population selection on changes in mRNA expression by assessing two wild populations from different river systems. Further studies by Debes et al. illustrated the effect of wild rearing environment on differential mRNA expression when either wild population was paired to domesticated populations. The present study aimed to extend previous experiments by our group and others by examining differences that occur within a specific strain reared in the same environment, with one wild group a year older than the other. Domesticated populations were matched to wild-type fish of either the same age or the same size (year older) in order to distinguish alterations in mRNA expression that have arisen due to differences in fish developmental stage rather than due to domestication.
Significant differences in mRNA expression were noted for age-matched relative to size-matched wild-type trout, with 0.68 per cent of all detected mRNAs differing greater than 2-fold (2.4 per cent for all differentially detected mRNAs). When domesticated trout were compared to wild-type trout groups, unique differences in mRNA levels were noted. This comparison reveals how developmental stage and/or age, can alone cause differential mRNA levels, independent of effects arising from domestication. Further confirmation of these findings was found upon analysis of the effect of genotype on mRNA regulation comparing parental to hybrid groups. Unique mRNA expression patterns were observed between domesticated and either age-matched or size-matched wild-type trout, consistent with a clear effect of fish development stage on mRNA levels. The biological pathways influenced by these mRNAs in both groups of wild-type trout also differed, primarily with respect to transport, response to stimulus, and, most strongly, cell/tissue structure and development, and lipid metabolism. The manner in which wilds differed in terms of physiology strongly supports the case of difference due to age, development, and life history. These results suggest that caution should be applied in interpreting data where only one control group (age- or size-matched) is selected in experiments comparing fish with different growth rates.
Conclusion
The present study has shown that considerable differences in genetics and physiology are associated with strains of domesticated and wild-type rainbow trout. Assessment of genotype effects demonstrated mainly additive and Drecessive mRNA regulation in hybrid progeny. To better understand the consequences of hybridization (and effects on phenotype and risks of introgression of domesticated strains into native populations), further study would be beneficial, including assessing additional strains and species, allelic variation among individuals within strains, and second generation hybrid crosses to identify specific quantitative trait loci influencing morphology, physiology (e.g. growth), and mRNA levels (eQTL analysis). The mRNAs within the D-recessive class may be of particular interest in this regard as we predict that these genomic blocks may show highly significant eQTL associations with growth. The present study revealed the importance of assessing the effect of developmental stage on differential mRNA levels between fastgrowing domesticated and slow-growing wild-type groups. Differences in mRNA levels (from that seen in domesticated trout) were more prominent for age-matched as opposed to size-matched wild-type controls, suggesting that matching groups by size (e.g. developmental stage) may provide more biologically meaningful data indicative of genetic differences between strains.
November 2013




