Introduction
Amoebic gill disease (AGD) occurs in mariculture of salmonids worldwide, but has caused particular problems to the Atlantic salmon (Salmo salar) farming industry in Tasmania. There have recently been widespread and sustained outbreaks of AGD also in Ireland and Scotland (Mitchell & Anderson inAdams et al., 2012). In fish with AGD, amoebae cover the gill epithelium and cause hyperplasia, hypertrophy, lamellar fusion and cavitations ( Adams and Nowak, 2001 and Adams and Nowak, 2003). Affected gill regions often appear as visible, pale patches (Adams et al., 2004). Several members of the genus Paramoeba (syn. Neoparamoeba see Feehan et al., 2013) have been isolated or detected in marine fish with AGD, P. pemaquidensis, P. branchiphila and P. perurans. In-situ hybridization studies showed that of these, only P. perurans is directly associated with the AGD lesions (Young et al., 2007). Paramoeba perurans has recently been cultured, and proven to be the causative organism of AGD through challenge experiments (Crosbie et al., 2012).
Using in-situ hybridisation, it have been shown that P. perurans was also the agent of AGD in seawater reared Atlantic salmon from Washington (USA), Chile, Ireland and Scotland, in farmed Chinook salmon (Oncorhynchus tshawytscha) from New Zealand, rainbow trout (O. mykiss) from Tasmania and in turbot (Scophthalmus maximus) from Spain ( Bustos et al., 2011 and Young et al., 2008b). Using PCR-techniques, this amoeba has also been detected in farmed Atlantic salmon with AGD lesions in Norway (Steinum et al., 2008), and in farmed ayu (Plecoglossus altivelis) in Japan (Crosbie et al., 2010). AGD has also been observed in wild blue warehou (Seriolella brama), farmed sea bass (Dicentrarchus labrax) and farmed sharpsnout seabream (Diplodus puntazzo), but whether P. perurans was present in these cases is unknown (see Feehan et al., 2013). Hence, apart from turbot and ayu, the records of P. perurans infections are from salmonids.
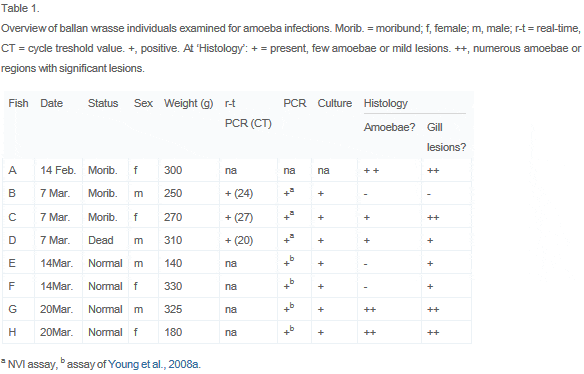
Ballan wrasse (Labrus bergylta) and other Labridae are used as cleanerfish in Atlantic salmon farming, for the control of sea lice infections ( Kvenseth, 1996 and Skiftesvik et al., 2013). During rearing of ballan wrasse broodstock, used for experimental production of fry, moderate mortality was experienced in connection with spawning. The fish were healthy, apart for a few moribund individuals. A health check revealed pale patches on the gills, and gill lesions associated with amoeba-like organisms were observed by histopathological examination. The presence of P. perurans was subsequently verified by molecular methods. The present paper reports this first observation of amoebal infection and AGD lesions in ballan wrasse (Table 1).
Material and methods
Ballan wrasse is experimentally cultured at the Institute of Marine Research (IMR) research station at Austevoll, near Bergen, Norway. The broodstock studied here was kept in two round 7000 l tanks with artificial shelters (plastic ‘kelp’) (Fig. 1A). The fish in tank A was a mix of broodstock of hatchery origin and wild caught (collected locally 2010–11), tank B contained only broodstock of hatchery origin, but a wild caught male was briefly introduced in the tank in 2012. The water is pumped from 165 m depth, filtered through a sandfilter (ca. 30 μm) and then through 20 μm (Arkal-filters). Fish density was low, only about 60 specimens per tank. The inlet water was heated and kept at 11.0–11.5 °C in the tanks, and the fish was light manipulated in order to stimulate spawning 10 weeks prior to the natural cycle. Amoebae were first noted histologically in a fish sampled 14th February 2013. Additional samples were then collected 7th March (N = 3) (histology, real-time PCR, PCR and culturing), 14 March (N = 2, PCR, histology) and 20th March (N = 2, PCR, histology, culturing). The studied fish weighed 140–330 g. For histology, samples were fixed in phosphate buffered 10 per cent formalin, embedded in paraffin according to standard protocols and 3 μm sections were stained with haematoxylin and eosin or haematoxylin-erythrosine-saffron (HES). Gill smears were stained with Giemsa or HES. Measurements on amoebae are given as range with mean in parenthesis. The mean diameter of each amoeba was calculated from their maximum diameter and the diameter perpendicular to that axis.
Molecular diagnosis was done both at the IMR and at the National Veterinary Institute (NVI). DNA was extracted from ethanol-conserved gill pieces or cultured amoebae using the DNeasy® Tissue Kit protocol for animal tissues or the QuiaQube robot (Qiagen). For conventional PCR targeting Paramoeba-18S rDNA the primers of Young et al. (2008a) and the following forward/reverse primer combinations were used (IMR): Erib1/NeoEK-R2, NeoEK-F1/Erib10 and NeoEK-F2/Erib10. These primers amplify fragments of 633, 1032, 1137, 1103 bp respectively. The novel primers has sequences 5′-CAAGAATTTCACCTCTGACAATGA-3′ (NeoEK-R2), 5′-ATTAGAGTGTTCAAGGCAAGCA-3′ (NeoEK-F1) and 5′-TCTGCAATGAATACTATTAGCAT-3′ (NeoEK-F2), while the sequence of the Erib primers are given in Barta et al. (1997). The PCR amplifications were performed in a total volume of 50 μl using 2 μl of template DNA and a reaction mixture consisting of 10 μl 5x PCR buffer, 3 μl 25 mM MgCl2,5 μl 10 mM dNTP, 2 μl (10 mM) of the reverse and forward primer, 2 U of thermostable DNA polymerase (GoTaq) and 26 μl dH2O. The PCR conditions were as described in Køie et al. (2008). An annealing temperature of 52 °C was used in these PCRs. Conventional PCR at NVI was performed using illustra PuReTaq Ready-To-Go PCR Beads (GeHealthcare) using the primers Neopara_F1_HH (5′-CTACTTGGATAACCGTGGTA-3′/Neopara_R1_HH (5′ CCTTGAACACTCTAATTTACTC-3′) and PeruFWD_HH (5′-GTTCTTTCGRGAGCTGGGAG-3′)/Neopara_R1_HH. The PeruFWD_HH primer was modified from Fringuelli et al. (2012) while the other primers were designed to amplify a fragment of approximately 750 bp of P. perurans including variable regions. An annealing temperature of 50 °C was used in both these PCR reactions.
The PCR products were cleaned with ExoSAP-IT® (Affymetrix Inc.) and then sequenced using a BigDye® Terminator v3.1 Cycle Sequencing Kit. The sequencing (both sense and anti-sense strands) was done using the amplification primers. The sequence data were assembled with the Vector NTI 11 software (Invitrogen) and subjected to BlastN searches in GenBank for similar sequences.
Real-time PCR was done at NVI with the assay and set-up of Fringuelli et al. (2012).
Amoebae were isolated for culturing as described by Morrison et al. (2004) using distilled water, but also by using autoclaved seawater and by swabbing gill pieces directly onto the agar. Cultures were established as described by Crosbie et al. (2012), and subcultured after 10 to 14 days (4 passages).
Results
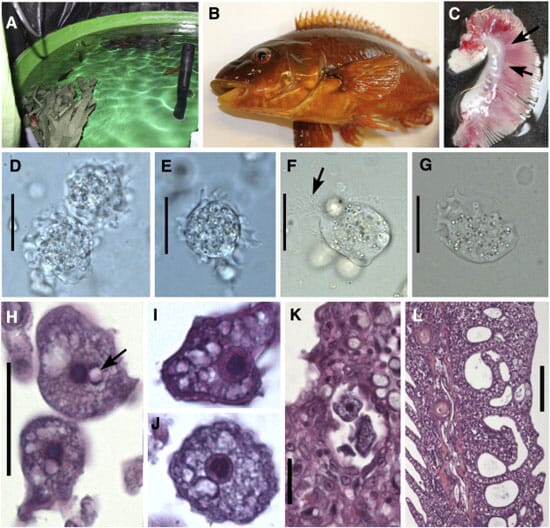
Both moribund and apparently healthy ballan wrasse (Fig. 1B) showed pale patches on the gills, most commonly located basally on the filaments (Fig. 1C). Wet-preparations from these areas showed numerous amoebae with slowly moving pseudopodia, measuring 17.3–32.5 (23.8) μm (N = 35) in mean diameter (Fig. 1D,E). Amoebae in stained air dried smears measured 16.9–34.2 (23.1) μm (N = 64) in mean diameter.
Yeast-malt extract agar cultures showed extensive amoebal growth independent of isolation methods employed. Detached spherical amoebae measured 22.4–28.5 μm in diameter.
Molecular analyses
Following the first incidental histological detection (14th February), new samples were obtained for molecular detection and identification of the amoebae. All the additional seven fish examined were positive for P. perurans with real-time PCR (7 March samples) or diagnostic PCR (14th and 20th March samples). Using primers that amplify all known Paramoeba spp. from fish, partial SSU rDNA sequences of P. perurans were obtained both from cultures and directly from infected gill tissue (14 and 20 March samples). Assembled sequences (2028 nt) were deposited in GenBank with the Accession numbers KF146711-3. Further sequences were obtained from the 7 March samples, a consensus sequence (692 bp) was identical to KF146712, with reference to that sequence covering positions 150–841.
Sequence identity was 99.3–99.9 per cent to P. perurans sequences from Salmo salar in Norway, 99.7–99.8 per cent to a sequence from S. salar in Chile, and 98.7–99.4 per cent to sequences from S. salar in Tasmania (GenBank accession numbers in: Bustos et al., 2011, Nylund et al., 2008, Steinum et al., 2008 and Young et al., 2007). Ambiguous signals, detected at 10 positions, always reflected variation previously reported in P. perurans.
Histology
The histopathological examination (8 fish) of the most affected gills revealed pronounced changes dominated by hyperplasia of poorly differentiated epithelium and fusion of lamellae, forming interlamellar spaces when fused at the apical end (Fig. 1L). Focal recruitment of mucus cells was observed, and affected areas contained eosinophilic granular cells, rodlet cells and chloride cells as well as some leucocyte infiltration. On top of hyperplastic tissue and in interlamellar spaces there were mucus, cell-debris and free or attached amoebae. In some fish the lesions observed were mild to moderate and located at the base of the filaments. No amoebae were detected in unaffected parts of the gills.
The amoebae were granular, often with clear vacuoles, and contained a nucleus with a large dark karyosome. The karyosome usually was round (Fig. 1H,I), but occasionally appeared horseshoe-shaped when sectioned (Fig. 1J). The nucleus was accompanied by one, rarely two visible juxtanuclear parasomes (Fig. 1H,J). These were usually pale in the center and basophilic at the periphery, particular at two poles. When sectioned through the nucleus, the amoebae measured 13.6–24.1 (17.4) μm (N = 61) in mean diameter, but reached 37 μm in length.
Some of the ballan wrasse were also infected with epitheliocysts (in chloride cells), Loma sp. xenomas (Microsporidia) and the monogenean Microcotyle donavani (two fish) in the gills.
Discussion
Previous findings of P. perurans are from farmed fish with AGD. Virtually nothing is known on the ecology of this amoeba, but it is probably an opportunistic parasite with a reservoir in the environment. We do know that P. perurans appears to be cosmopolitian in distribution, and also that it is able to infect unrelated fish species in mariculture; i.e. showing low host specificity. Its occurrence as an ectoparasite seems usually to be associated by localized lesions, suggesting virulence, although moderate numbers of patchy lesions on the gills need not seriously affect the hosts. Although gill lesions and histological changes characteristic of AGD were detected in the studied ballan wrasse, we found these in both clinically healthy and in moribund fish. Impaired spawning (egg release) may have caused mortality among females in studied wrasse population; there were no apparent signs of respiratory distress. However, moribund males were also observed, apparently with light P. perurans infections. Agonistic behaviour may be a primary cause for mortality among the males. A possible source of the amoebae in both the studied tanks is wild caught ballan wrasse individuals, which had been collected 2–3 years previously. This suggests that P. perurans infections may have persisted in one of the tank populations for years, the other tank with broodstock of hatchery origin may have obtained amoebae from a single male introduced in 2012. The inlet water was obtained from a considerable depth (165 m) and filtered. However, filtering at 20 μm does not exclude the deepwater as a possible source of the amoebae.
The SSU rDNA sequences of amoebae obtained from ballan wrasse or cultures identify the dominant amoeba as P. perurans. Since unrelated fish species such as ballan wrasse and Atlantic salmon are both colonized by P. perurans, this amoeba likely has the potential to infect a wide range of fish species. Whether P. perurans infections are common on fish in nature is presently unknown, farmed fish may be stressed, immuno-compromised or otherwise weakened and thus more susceptible.
To our knowledge the present observations represent the first finding of P. perurans infections in ballan wrasse, and the first case of AGD in a wrasse (Labridae). However, it does not represent the first case of amoeba infections in wrasse. Chatton, 1909 and Chatton, 1910 described a case of amoebosis in tank-reared Symphodus tinca and S. melops from Banyuls-sur-Mer, France. The amoebae occurred in large amounts in the gills of the wrasse, and caused respiratory distress. They measured 12–30 μm in diameter, but usually about 20 μm in sections, with a 3 μm nucleus. Chatton's images of Amoeba mucicola from sections does not show any clear parasomes. Still it appears possible that this otherwise similar amoeba is a species of Paramoeba, and Amoeba mucicola should be redescribed from its Mediterranean labrid hosts and characterized using molecular methods.
Since ballan wrasse is used as cleanerfish in salmon farms, the introduction of these to the pens may represent a source of P. perurans infections in salmon. Hence, the role of wild ballan wrasse and other cleanerfish species as hosts for this amoeba needs to be studied. Still, more likely P. perurans reservoirs are the salmon farms themselves and free amoebae in the water. Clearly, research is urgently needed on the biology and epizootiology of this recently described and therefore poorly known amoeba.
February 2014




